
"I have used a wide variety of secondaries and Jackson ImmunoResearch has consistently been the best. The fluorophores are bright and stable and their selective (x reactivity removed) secondaries have always shown species specificity in multiple labeling."
Janet Duerr, Ohio UniversityRating: 5.0
Protein tags are regularly used in molecular biology to enable the confirmation, quantification, and purification of fusion proteins. They range from whole functional proteins such as GFP, domains such as an Fc from an immunoglobulin, or short peptides like His-Tags. Here, we demonstrate two experimental techniques, Western blot, and ELISA, to analyze transfection efficiency by confirming and quantifying the expression of a GFP fusion protein using our NEW Anti-GFP antibody.

Epitope tags are regularly used in the generation of fusion proteins. Tags enable the detection and/or the purification of proteins of interest from expression media. The tags can also improve solubility, improve protein expression by influencing correct protein maturation, and be used to characterize protein interactions. Epitope tags can be added to a protein to confer additional function without changing the structure of the protein of interest. The addition of cleavage sites allows tags to be removed from the protein during purification stages, leaving a functionally competent protein for downstream applications. Tags may be full-length proteins such as Green Fluorescent Protein (GFP), specific domains such as an Fc domain from an immunoglobulin, or short peptides like a His-tag.
GFP tags are popular reporter tags. They are well-characterized proteins that can be readily expressed as functional fusion proteins. GFP tags can also help study protein-protein interactions when paired with other fluorescent molecules.
The expression of fusion proteins may be performed in a variety of expression systems, including mammalian, bacterial, and yeast. Two applications that are commonly used to confirm expression and assess transfection efficiency are Western blotting and ELISA.
These techniques can be performed using antibodies specific to the protein of interest or antibodies directed against the epitope tags incorporated into the proteins. The advantage of using antibodies specific to a protein tag is that they are often well-characterized and easily available, making them more reliable and cheaper. Targeting tandem epitope tags, for example a GFP-tag used in combination with a His-tag, allows researchers to compare expression characteristics of different constructs.
The following case study demonstrates the qualification and quantification of the expression of a GFP fusion protein by Western blotting and ELISA using Jackson ImmunoResearch's NEW Anti-GFP Antibody.
In this case study, transfection efficiency was assessed using Western blot and ELISA. HEK 293T cells were transiently transfected with EGFP (see experimental setup) by titrating the transfection reagent, PEI, and DNA. The influence of these variables on protein expression was compared using an HRP-conjugated Rabbit Anti-GFP antibody from Jackson ImmunoResearch.
HEK 293T Cells were grown in T75 flasks at 37°C, 5% CO2 to 100% confluency. Then, cells were subcultured into nine T25 flasks followed by incubation at 37°C, 5% CO2 for 2 hours.
Post incubation, cells were transfected with PEI (Tocris) and DNA (pTwist-CMV-Hygro encoding EGFP, Twist biosciences) in the volumes as described below.
| T25 flask | PEI (μg/cm2) | DNA (ng/cm2) |
|---|---|---|
| 1 | 0 | 0 |
| 2 | 1 | 0 |
| 3 | 1 | 50 |
| 4 | 1 | 100 |
| 5 | 1 | 200 |
| 6 | 2 | 0 |
| 7 | 2 | 50 |
| 8 | 2 | 100 |
| 9 | 2 | 200 |
24 hours post-transfection, cells were washed with DPBS, followed by the addition of 5 ml of DPBS. Cells were then harvested using a cell scraper and centrifuged at 200xG for 10 mins. Cell pellets were lysed in 220 μL of MPER Mammalian Protein Extraction Reagent (Thermo Fisher), and total protein concentration was measured at A280 nm by Nanodrop.
Western blotting is a quick and sensitive method that is regularly used to confirm the expression of target proteins. Small volumes of cell lysate from practically any expression system can be quickly analyzed with crude preparation. Proteins of interest can be detected using an antibody against the target protein or, more commonly, the epitope tag used to generate the fusion protein. By targeting the fusion protein, one antibody can be used to screen limitless constructs, which can help improve screening efficiency and economy. Using a conjugated secondary antibody for indirect detection can also improve the sensitivity of the western blot by amplifying signal.
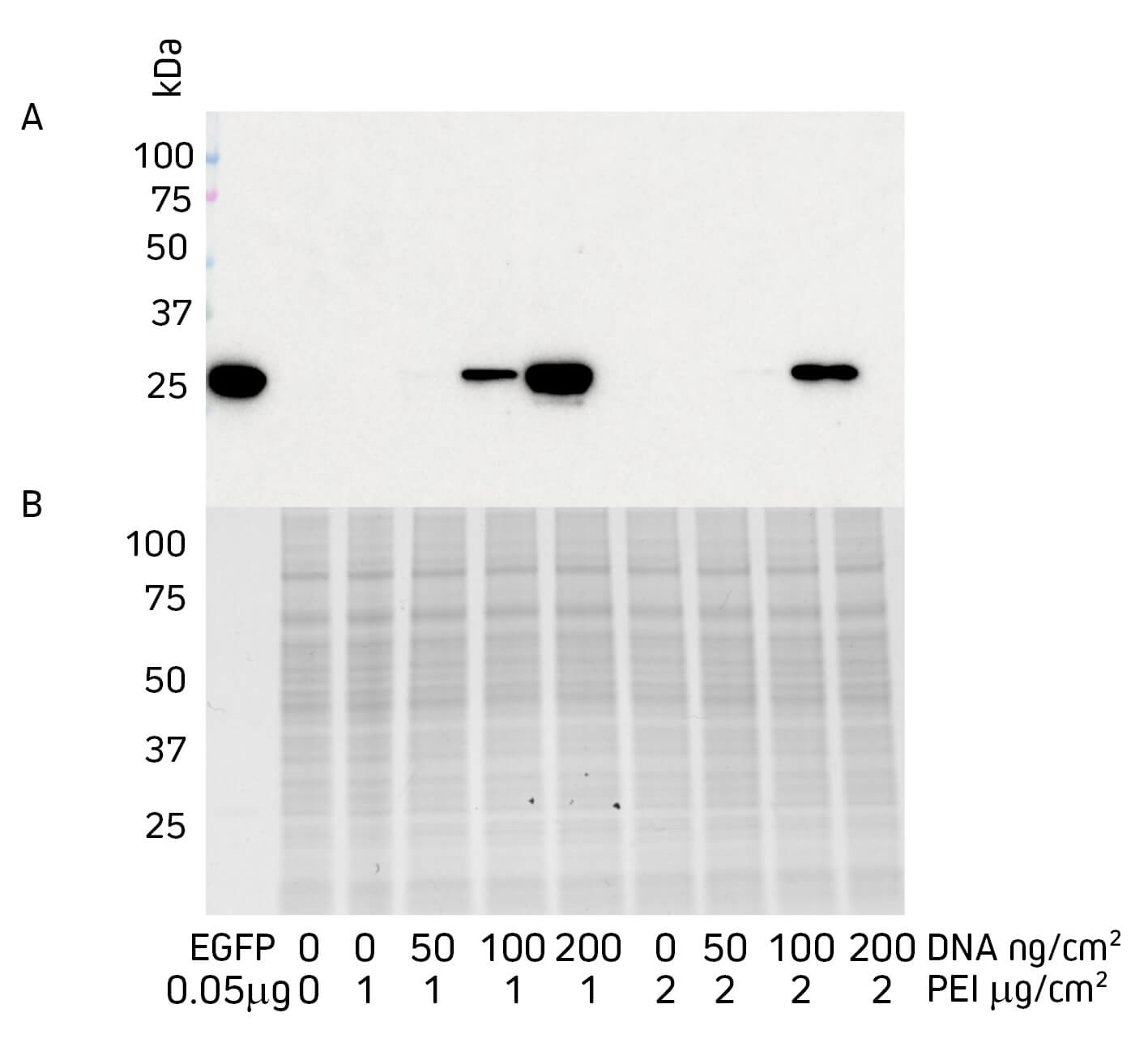
Samples were denatured and reduced (boiling at 100°C for 10 mins in 5% BME+Laemmli buffer) and loaded into precast gradient gels 4-15%, 12-well comb (Biorad, 4561085). Purified EGFP was loaded into the control lane at 0.05 μg/well. Samples were loaded at 20 μg/well of total protein as measured by A280 nm. Samples were run at 150 V for 50 mins.
Figure 1 shows how changing the quantity of DNA and the transfection reagent PEI can influence protein expression. In this experiment, it can be seen that increasing the amount of DNA improved protein expression. However, increasing PEI appeared to decrease expression and negate the effect of adding more DNA, which is consistent with previous assays that suggest PEI is toxic to the cell above certain concentrations (Lewis et al., 2013).
The comparison of Western blot to Coomassie-stained SDS PAGE gel shows the sensitivity of the blotting process, where the seemingly absent protein in the SDS PAGE gel is clearly shown as abundant signal by Western blot. The power of this application is that very small expression experiments can be performed, reducing the pull on resources whilst allowing a number of constructs and/or expression conditions to be interrogated before scaling up expression.
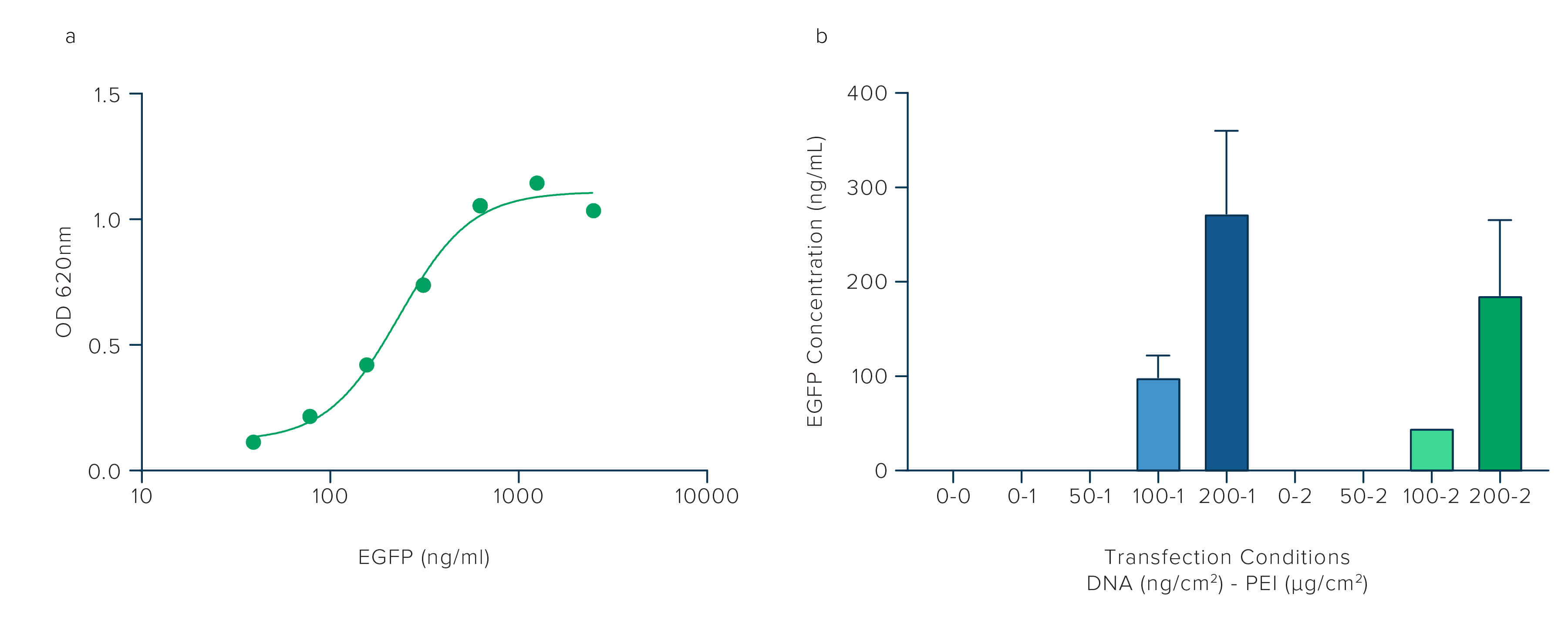
ELISA can be used to confirm and quantitate protein expression. In the experiment below, we target the EGFP using JIR's Anti-GFP antibody to compare the transfection efficiency achieved when the two variables, PEI and DNA, are altered. The ELISA allows the quantification of protein expression under each condition. Signals from the samples are compared to a standard curve generated using purified EGFP protein. The sample results are then interpolated from the standard curve to give an estimated concentration.
96 well microplate was coated with Rabbit Anti-GFP (300-005-245) at 10 μg/mL diluted in 0.1M NaHCO3/NaCO3, 0.05% Sodium Azide, pH 9.6. Following overnight incubation, the plate was washed 3X in PBS/T and blocked with 1% BSA in PBS/T for 2 hours.
The purified EGFP standard curve was applied to column one, loaded at 250 ng/ml in the first well, and serially diluted 1:2 down the subsequent six wells, with a blank (PBS/T) in the final well.
Cell lysates were diluted to 1 mg/mL and loaded at 1:2.5 dilution in the first well of columns 2 through 10, then serially diluted 1:2 down six wells, followed by blank (PBS/T) in the final well.
Protein concentrations were calculated by interpolating sample signals against EGFP standard curve using Graphpad Prism software.
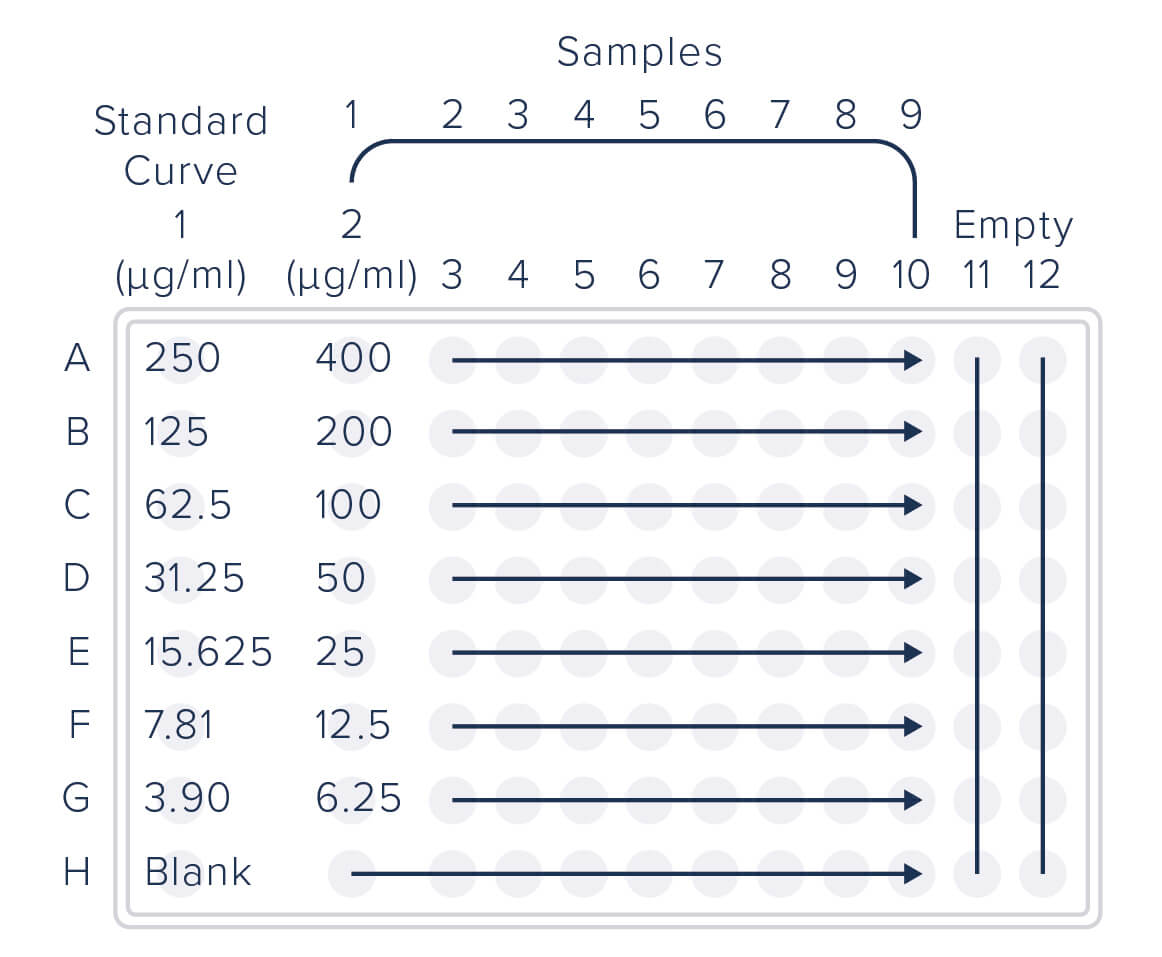
Figure 2 shows how protein expression can be quantified by ELISA using Jackson ImmunoResearch Rabbit Anti-GFP antibody in combination with a purified EGFP standard curve. Figure 2b corroborates the findings of the western blot in Figure 1, adding more tangible quantitation that increasing the amount of DNA used in the transfection improves protein expression, whereas increasing PEI decreases expression and negates the effect of adding more DNA. ELISA, as a technique, offers sensitive, accurate, and quantifiable data. Using an antibody that targets the GFP tag rather than the target protein avoids the need to purify the target protein to generate a standard curve to calibrate protein concentration against.